Numerical simulation of structure and properties of polyphilic molecules in phospholipid membranes
The aim of this project is the simulation and the computational characterization of polyphilic molecules with lipid bilayers. From a methodological point of view, molecular dynamics simulations based on force fields are combined with ab-initio (quantum chemical) molecular dynamics simulations and spectroscopic calculations.
Several types of polyphilic molecules (linear, X- and T-shaped polyphiles) are studied in interaction with phospholipid bilayer membranes. Besides hydrophilic and hydrophobic parts, the molecules contain fluorophilic segments. For the linear and X-shaped molecules, the orientation of clusters of these molecules inside and across the phospholipid membranes will be of interest, while the T-shaped polyphiles will be simulated in the framework of twodimensional membrane segments, where they are expected to stabilize the bicellular structure. Besides the morphological superstructure of these two-component-systems, a particular focus is the function and localization of the fluorophilic groups.
The aim of the molecular dynamics simulations is to obtain an understanding of the structural driving forces that govern the supramolecular selforganization process in these systems, but also to elucidate the way in which the polyphile molecules (and in particular the fluorinated segments) modify the properties of the membrane as such. One the one hand, this is related to the interaction of the fluorophile parts with the polar membrane-water interface, and on the other hand, it targets the properties of the membrane as an entity, in view of the mobility and diffusion properties of the lipid molecules and the permeability of the membrane with respect to water molecules. The results of the molecular dynamics simulations are compared with experimental data from NMR and microscopy, in order to verify the output of the simulations, but also in order to interpret the experimental findings on a molecular level.
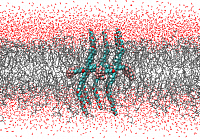
Simulated cluster of X-shaped bolaamphiphiles in a phospholipid bilayer
